GWAS4D Quick Tutorial
GWAS4D is best accessed using the latest version of Chrome/Firefox/Safari web browser. Not all functions are available with Internet Explorer, due to a lack of SVG and HTML5 features.
Job submission
Input profile (hg19/GRCh37)
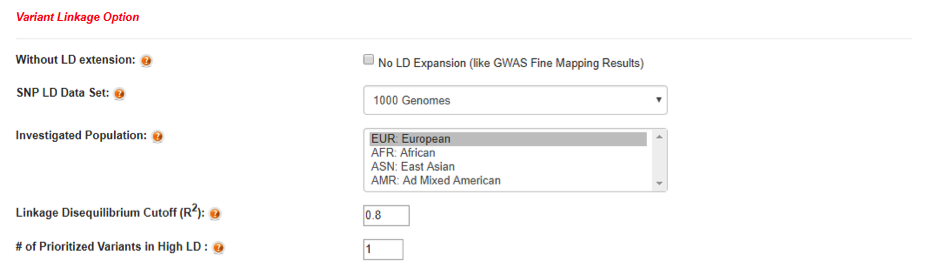
GWAS4D accepts four different commonly used GWAS summary statistics result formats in hg19/GRCh37, and it supports either plain text input or uploaded file:
- 1) Variants Coordinates: Chr, Pos, [P-value]
- 2) VCF-like Map: Chr, Pos, SNPID, Ref, Alt, [P-value]
- 3) Single SNP ID: dbSNPID, [P-value]
- 4) Plink-like Map: Chr, dbSNPID, Pos, [P-value]
Note: The delimiter should be TAB or comma. User can either click "example" text or download "example input files" for GWAS4D testing.
GWAS4D can also filter out the non-significant GWAS signals. Also, it allows inputting variant list without GWAS P-value (all of variants will be considered in this case). To reduce the computational load of backend server, GWAS4D now supports no more than 10k input significant variants under "without LD extension" mode, and 2.5k input significant variants under "LD extension" mode.
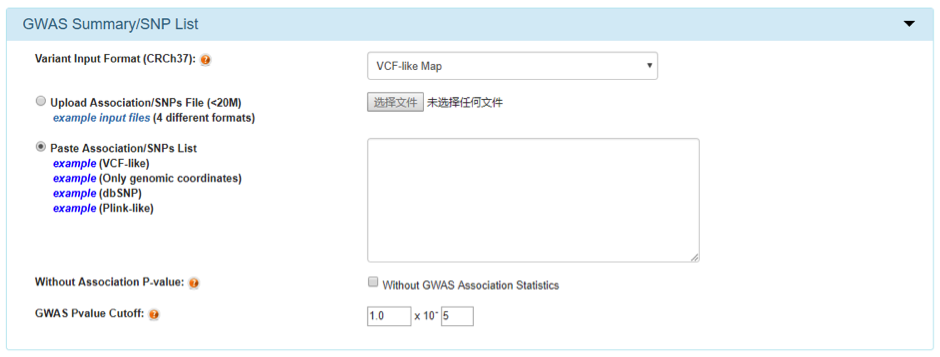
Variant Linkage Option
GWAS4D allows adjusting LD cutoff or disabling LD extension function. GWAS4D supports LD structure of many subpopulations, which were calculated from 1000 Genomes project phase 1v3 and HapMap I+II+III genotype.
Also, it supports defining the number of independent variants in highly linked group, which means GWAS4D outputs given number of variants with the top largest combined regulatory probabilities in each high LD proxy.
Note: We suggest users to disable LD expansion function when input GWAS signals are from GWAS fine-mapped credible set or conditional analysis.
Tissue/Cell Type Option
GWAS4D adopts our cepip algorithm (Li MJ et.al. Genome Biology. 2017 16;18(1):52) to prioritize context-specific regulatory variant, which supports Roadmap epigenomes for 127 tissues/cell types. To acquire the details regarding to these 127 tissues/cell types, please visit http://egg2.wustl.edu/roadmap/web_portal/.
User can also upload own chromatin ChIP-seq/DHS/ATAC peaks for particular tissue/cell type.
For unknown GWAS disease/trait-associated tissue/cell type, GWAS4D can estimate likely relevant tissue/cell type in 127 Roadmap tissues/cell types by checking corresponding option.

Note: User-defined epigenome file should comply with ENCODE narrowPeak-like format as follows or download example (delimiter should be TAB):
chrom chromStart chromEnd name score strand signalValue pValue qValue peak markName
chr1 10051 10552 Rank_15424 135 . 5.43713 13.53831 10.60377 200 H3K27ac
The markName of epigenome is compulsory field in the last column, including DNase, H3K27ac, H3K27me3, H3K36me3, H3K4me1, H3K4me2, H3K4me3, H3K79me2 and H3K9me3.
In addition, GWAS4D uniformly processes Hi-C data and generates significant interactions at 5kb resolution across 60 tissues/cell types from ENCODE project, 4D Nucleome project, as well as GEO database. It will automatically match tissue/cell type-specific epigenomes with Hi-C tissue/cell type. User can also upload own chromosome interactions region (5KB recommended) for particular tissue/cell type.
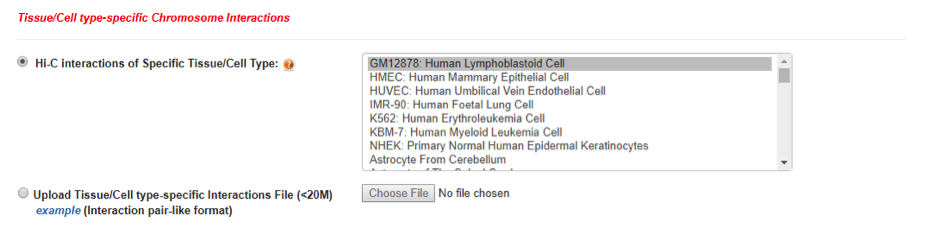
Note: User-defined chromosome interactions file should comply with following format or download example (delimiter should be TAB):
chrom_1 chromStart_1 chromEnd_1 chrom_2 chromStart_2 chromEnd_2 interaction_score
1 770000 775000 8 270000 275000 27.569397
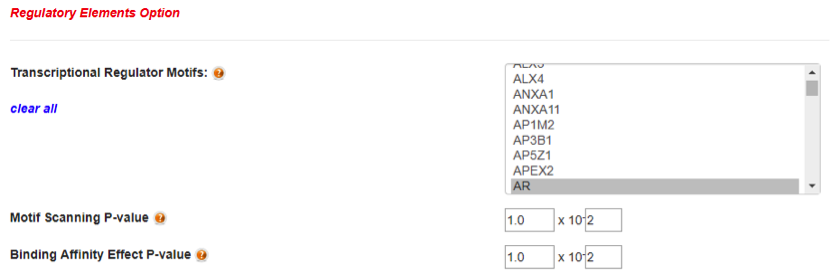
Regulatory Elements Option
GWAS4D integrates and refines motifs from eight public resources for 1480 transcriptional regulators. By default, GWAS4D only inspect whether regulatory variants alter 130 pre-selected transcriptional regulators. In addition, The P-value of significant motif scanning cutoff and the P-value of significant allele-altering binding affinity change can be adjusted.
Job tracking
Usually, GWAS4D can finish a task around 10mins. It supports job retrieving from three manners:
Fixed job URL (e.g. http://www.mulinlab.org/gwas4d/Z3dhczRkLTE1MTQ4MDMxODM)

Browser cookies in the Job menu
Send to email

Job result
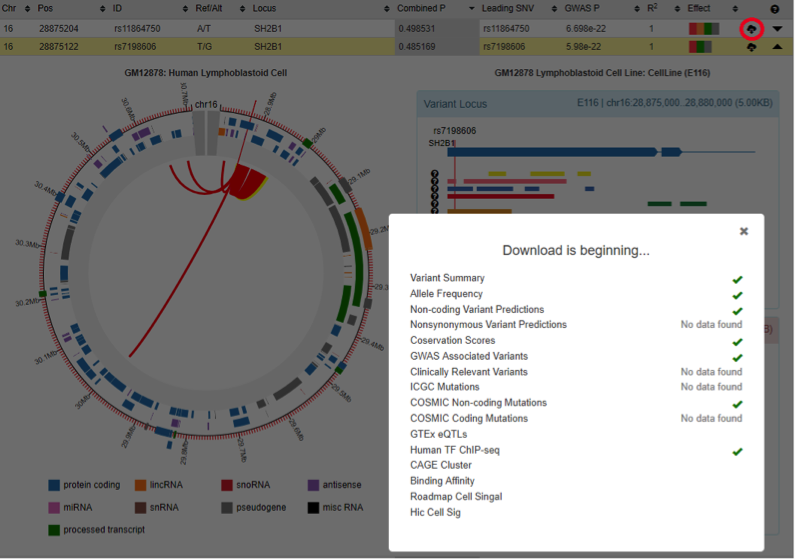
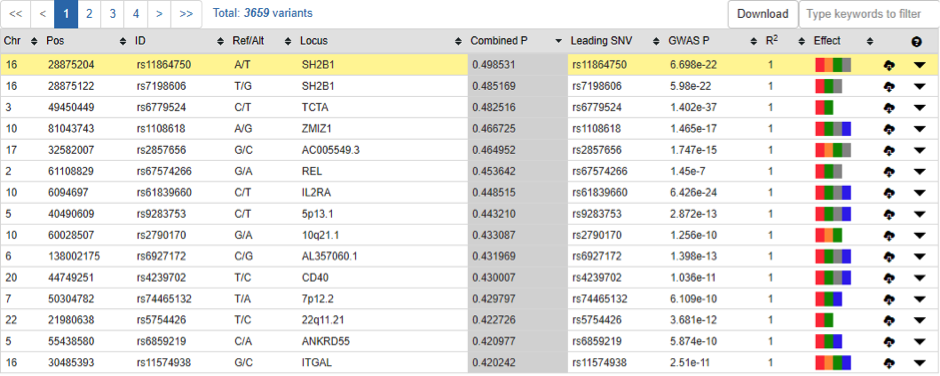
Regulatory variant prioritization table
By integrating latest multidimensional functional genomics resources and our regulatory prediction algorithm (cepip), GWAS4D extends LD variants and prioritizes them with combined regulatory probability. The variant prioritization table can be searched by typing key words, and can be downloaded.
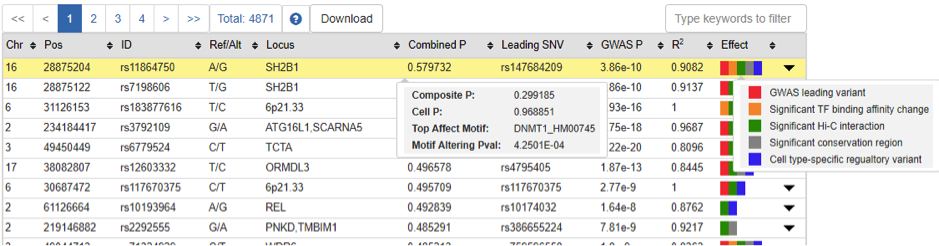
- 1) Combined P (recommended): combined regulatory probability calculated by our cepip, which jointly considers cell type-specific regulatory potential and cell type-free composite score (refer to PMID: 28302177)
- 2) Composite P: composite score calculated by our PRVCS (refer to PMID: 27273672)
- 3) Cell P: cell type-specific regulatory potential score calculated by our cepip (refer to PMID: 28302177)
- 4) Top Affected Motif: the most likely affected motif by alternative variant effect, including motif gain and motif loss
- 5) Motif Altering P-val: the significance of variant effect on binding affinity change of transcriptional regualtor
- 6) Effect: color schema that indicates whether this variant hit important regulatory signals, includes hitting to GWAS leading variant, significant TF binding affinity change, significant Hi-C interaction, significant conservation region, cell type-specific regulatory variant (top 10-quantiles)
- 7) Locus: map variant to GENCODE v27 for genic region, and map variant to UCSC cytoband for intergenic region
Variant-target inspection and visualization
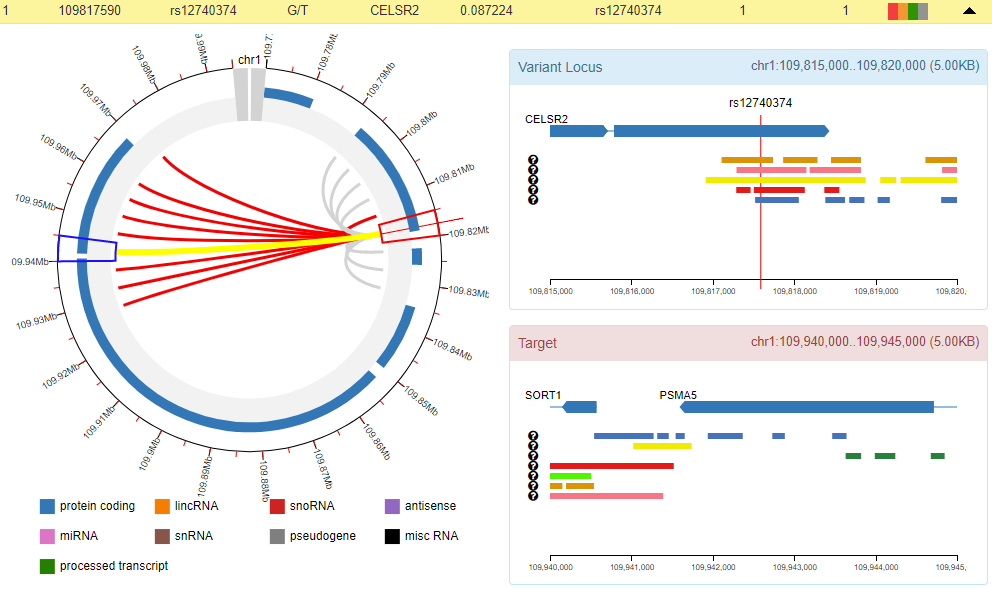
GWAS4D uniformly processes Hi-C data and generates significant interactions at 5kb resolution across many tissues/cell types of human organs and development stages. To facilitate the visualization of variant-target and associated chromatin states, we modified the js code of Capture HiC Plotter (chicp, refer to PMID: 27153610) and plotted top 10 significant Hi-C interactions at 5kb resolution.
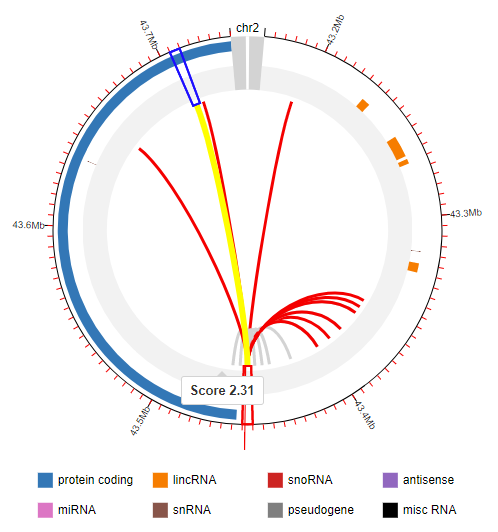
The red link is one of top 10 significant Hi-C interactions that called by Homer, and the grey link is one of top 10 contact signal that called by ICE. The genes and 5kb fragments can also be visualized in the plot.
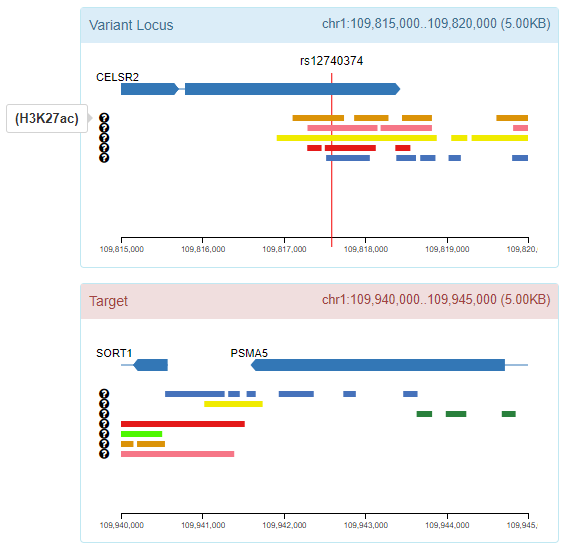
By clicking the interaction arc, GWAS4D plots the 9 associated chromatin states (map Hi-C tissue/cell type to the most relevant Roadmap epigenome) for variant locus (5kb) and target locus (5kb).
The interactive panel for circle plot and chromatin state plot:
Variant functional prediction and annotation
GWAS4D provides complete non-coding variant annotations and functional predictions:
-
1) Variants Information: variant genomic region, affected genes, variant allele frequency in different population

-
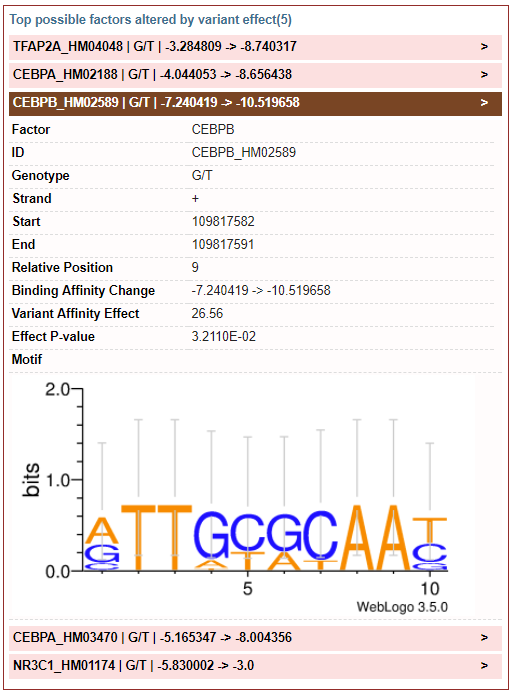
2) Binding Affinity: report top possible factors altered by variant effect (variant locus is labeled by relative position)
-
3) Functional Prediction: state-of-art non-coding variant functional scores, nonsynonymous mutation deleterious/pathogenic scores and base-wise conservation score
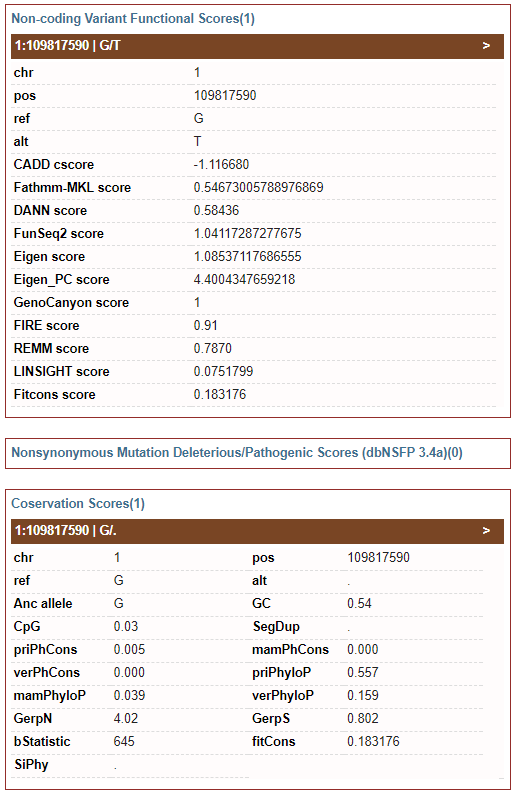
-
4) Disease Association: GWAS associations, clinically relevant variants and recurrent mutations
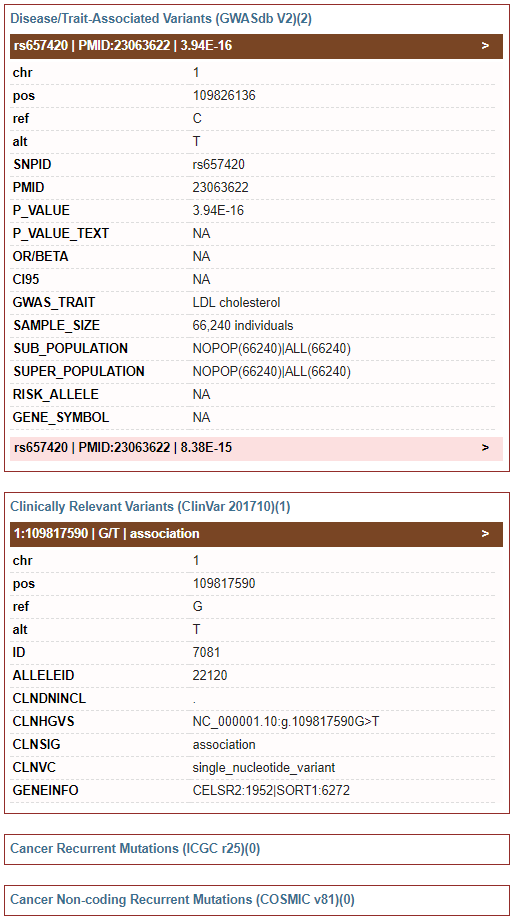
-
5) External Link: direct link to important regulatory variant annotation database
Download variant functional prediction and annotation
User can download all of functional predictions and annotations information for each prioritized variant by simply clicking a download icon in the variant prioritization table.