Summary
Allele-specific effects of cancer driver mutations influence patients’ drug responses and undermine the success of clinical trials. We reasoned that these effects will unbalance the distribution of each mutation across different cancers, as the result, the cancer preference can be used to distinguish the effects of them. Here, we developed a network-based framework to systematically measure cancer diversity for each driver mutation. Half driver genes harbor specific and pan-cancer mutations simultaneously, suggesting that functional heterogeneity of the mutations from even the same driver gene is very common. By associating the diversity spectrum with pharmacogenomic data, we demonstrated that specificity of mutations significantly influenced targeted drug responses. For most cases, the best drug responses were observed in patients harboring cancer-specific mutations. Moreover, we compared the diversity spectrums between primary and advanced tumors and found the diversity was generally increased in advanced tumors. Finally, a new protocol was designed to search for cancer genes based on the diversity spectrum. These results provide a foundation to optimize off-label targeted therapies and the design of clinical trials in the personal genomics era. All mutation annotations in this webportal are based on GRCh37.
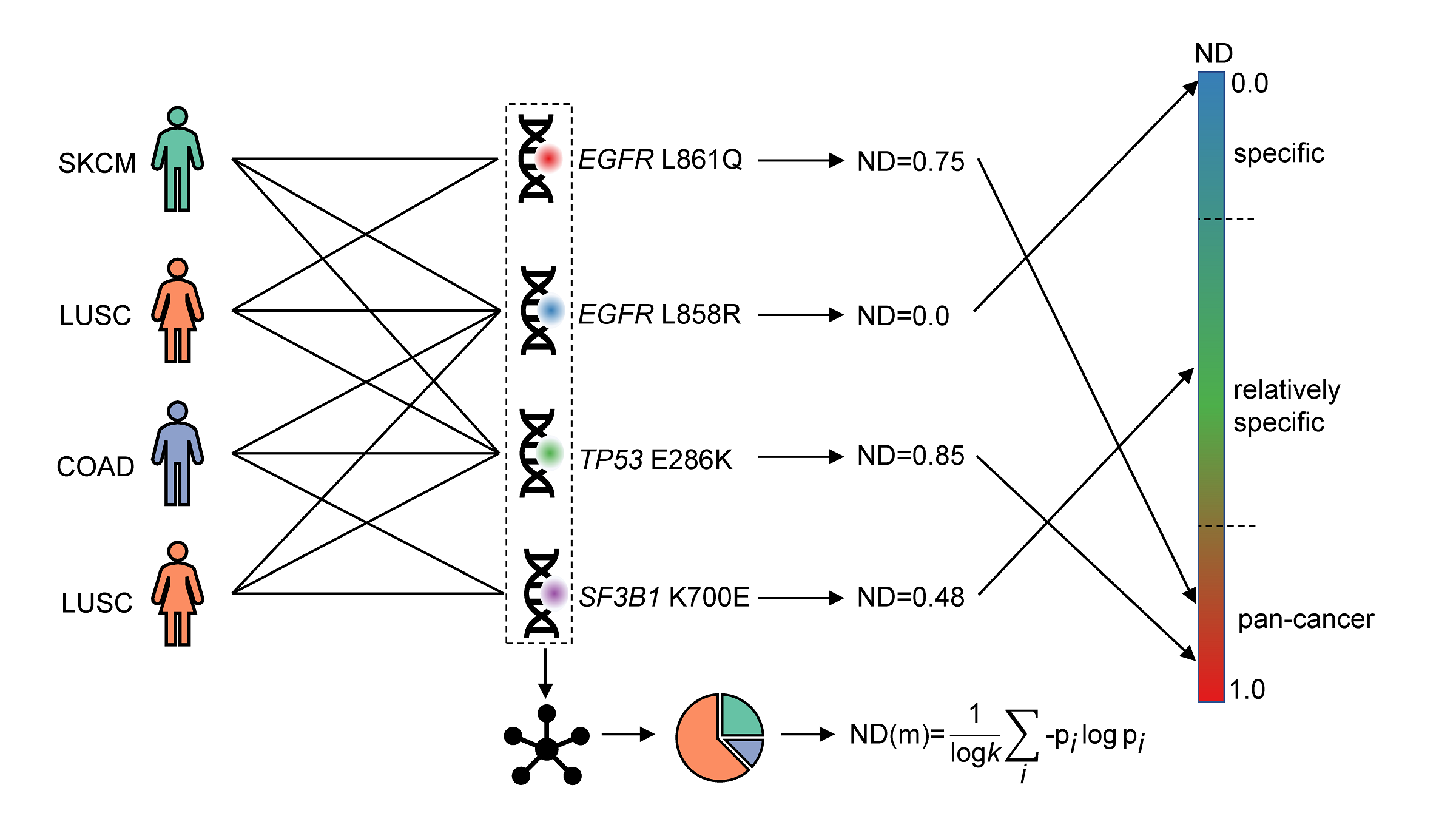
Patient-mutation Network
Patient-mutation network was exported in a Cytoscape Session (.cys) file
Reference
Dong et al. (2019). Diversity Spectrum Analysis Identifies Allele-Specific Effects of Cancer Driver Mutations. submitted
Contact
Firework was developed by mulinlab@Research Center of Basic Medical Sciences, Tianjin Medical University. If you have questions or suggestions to this platform, please contact us through mulin0424.li@gmail.com.
